Symmetry Coordinates
Symmetry coordinates, S, are specified by giving the
transformation matrix, U, between S and the
internal coordinates, R:
S = U R = U B Δx
The symmetry coordinates window, shown below for
C2H4, allows the display and editing of the
U matrix and (optionally) the force field in terms of
symmetry coordinates, F. To create a symmetry coordinates
object, right click on the Electronic
State object object and select "Add New...,
Symmetry Coords". To bring up the window right click on the
symmetry coordinates object and select "View...". If a symmetry
coordinates object is not present, or the U matrix is left
blank, an identity matrix is assumed for the U matrix.
Note that the current version of the program ignores the point
group set for the molecule as a whole, and the symmetry set for
the individual vibrational modes when doing the force field
calculation. This may be changed in future versions of the
program.

The Controls
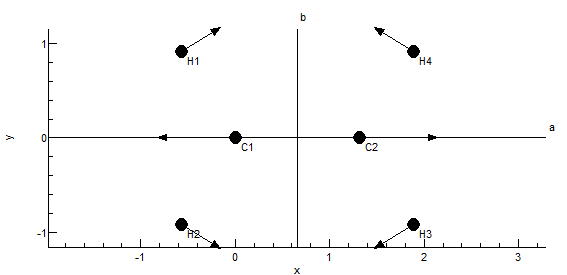
The plot at the bottom shows the current symmetry coordinate,
i.e. the selected row in the top left grid. The arrows indicate
how far each atom moves. The plot updates semi-automatically; use
"Operate, Check" to force an update.
+
 The "Plot Plane"
control allows the selection of the plane in which to plot the
molecule.
The "Plot Plane"
control allows the selection of the plane in which to plot the
molecule.
 The
magnification
selects the scale factor to use for the arrows.
The
magnification
selects the scale factor to use for the arrows.
 The status bar shows
the latest mouse position; draw a box with the mouse to measure
distance and angles.
The status bar shows
the latest mouse position; draw a box with the mouse to measure
distance and angles.

The operate button brings up a menu of possible operations:
Check
|
Check the contents of the top grids, and
update the plot to match
|
Apply
|
Check the contents of the top grids, and
display the details of the vibrational mode calculation in
the Log Window. This also forces
other objects to update.
|
| Show F Matrix |
Print the F matrix in the Log
Window calculated from the l matrix
elements and frequencies given in the vibrational mode objects.
The transformation is done by calculating the matrix L,
which relates the symmetry coordinates, S, and
normal coordinates, Q:
S = L Q
The L matrix is available from:
L = U B M-½
l
providing the internal coordinates are fully specified and
the F matrix can then be calculated from:
F = (L-1)T
λ L-1
Note that the U matrix is not necessarily required,
but if there are redundant internal coordinates L-1
can't be calculated. In this case an alternative
relationship can be used:
F = L λ
LT
provided that L LT= 1.
|
| Set F Matrix |
Clear the f matrix grid (top right) and set it
from the l matrix elements and frequencies
given in the vibrational mode
objects. See the entry above for details of the calculation. |
| Sort F Matrix by Value |
Sort the F matrix grid (top right) in descending
order of the first column. |
| Sort F Matrix by Mode |
Sort the F matrix grid (top right) by mode
numbers, with diagonal elements first. |
The Force Constant Matrix
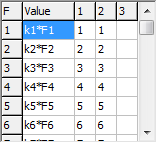
The grid at the top right allows the force field to be given in
terms of valence coordinates:
V = ½ ST F S = ½
Σij FijSiSj
The non zero elements of the
F matrix,
Fij are specified in the grid, with the
force constant in the first column, and
i and
j in
the next two columns. (The fourth column is to allow future
expansion to cubic and higher terms.) The force constants can be
given as simple numerical values, but can also be given as general
expressions involving symbolic values. This is typically more
useful as it allows, for example, relationships between
isotopologues to be added naturally. For the example shown here
the constants are expressed as the product of two factors, such as
k2*F2. The intent is that
k2 is a scaling
factor, as commonly used in interpreting
ab initio
calculations; here a different scaling factor is used for each
mode, but an obvious change would allow the same scaling factor to
be used for each mode. When reading the force constant matrix,
unknown variables are automatically created as parameters to the
current internal coordinates object, and they can then be edited
in the
Constants Window. If
dealing with more than one isotopologue, these variables are
better placed in a
variables object,
(with
Global set to
true) so the same value can be used in all species.
Symmetry Coordinates
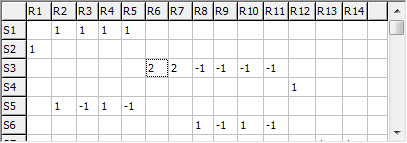
The grid at the top right allows the U matrix to be
specified; each row corresponds to a specific symmetry coordinate,
and the column the contribution from each internal valence (R) or
Cartesian (B) coordinate. As for the other matrices, either a simple
numerical value or a symbolic expression can be entered.
Internal Coordinates Settings
| Active |
Set true to use this symmetry coordinates object; there
must be no more than one active symmetry coordinates object
at a time.
|
| Normalize |
If true, the rows of the matrix given are normalized
before use in further calculations |
PreserveS
|
If true, the normal modes are in the same
order as the symmetry coordinates, using the largest
coefficients in the L matrix to align the two. This
is not guaranteed to work, particularly if there is
significant mixing between the symmetry coordinates.
If false, the normal modes are sorted by energy.
Note that the current version of the program ignores the
point group set for the molecule as a whole, and the
symmetry set for the individual vibrational modes when doing
the force field calculation. This may be changed in future
versions of the program.
|




