It is
normally more flexible to work with assigned lines in an independent
file for larger
fits as only limited editing is possible in the line list window. The
file is a text file normally with a .lin or .obs
extension and will accept a wide variety of formats and options on top
of the basic line list as displayed in the line window. For example, a "
stop" directive will cause the
rest of the file to be ignored, allowing fits to part of the data. For
full details see the section on
Line List
Input File Format. A custom text editor, the
Slave Editor, is distributed with
PGOPHER which has been
specially designed for editing these files. The procedure below
describes how to generate a
basic version of the file from the line list window.
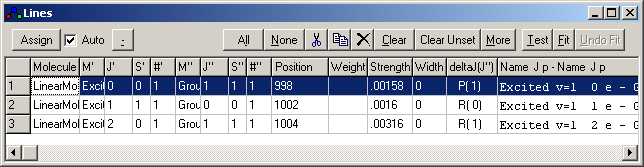
 . Note you can
adjust that the settings controlling the number of peak displayed and
assigned from the Line List Options Window.)
. Note you can
adjust that the settings controlling the number of peak displayed and
assigned from the Line List Options Window.)

 button in the log window will step
constants back one fit cycle for each press
button in the log window will step
constants back one fit cycle for each press button in the log window will step the constants
forward one fit cycle.
button in the log window will step the constants
forward one fit cycle.
 button and browse to
the file (or where you would like the file to be created).
This will put the file name in the observations
file name window. (The file name should typically end in .lin or .obs).
button and browse to
the file (or where you would like the file to be created).
This will put the file name in the observations
file name window. (The file name should typically end in .lin or .obs). . If the file does not exist you
will be asked if you want to create it. If the Slave Editor is set up correctly, this
should start.
. If the file does not exist you
will be asked if you want to create it. If the Slave Editor is set up correctly, this
should start. in the line list window) to put
your assignments onto the clipboard
in the line list window) to put
your assignments onto the clipboard in
the log window, rather than the line list window. If the Slave Editor is in use, then the contents
of all slave editors are saved before starting the fit. Note that is a
slave editor is unable to save a file that has been modified then the
fit will not start and the slave window will come to the front or flash.
in
the log window, rather than the line list window. If the Slave Editor is in use, then the contents
of all slave editors are saved before starting the fit. Note that is a
slave editor is unable to save a file that has been modified then the
fit will not start and the slave window will come to the front or flash.